Level and plasticity divergence in a common trans-regulatory environment
source("functions_for_figure_scripts.R")
library(ggExtra)
load("data_files/FinalDataframe3Disp.RData")
load("data_files/Cleaned_Counts.RData")
load("data_files/Cleaned_Counts_Allele.RData")
cor_thresh <- 0.8
eff_thresh <- log2(1.5)
# extreme cor and log2 fold change values to extract the most
# cis plasticity divergers and most trans level divergers for
# avg expression figures
cis_dyn_thresh <- -0.25
trans_lev_thresh <- log2(2)Level divergence tends to be in cis
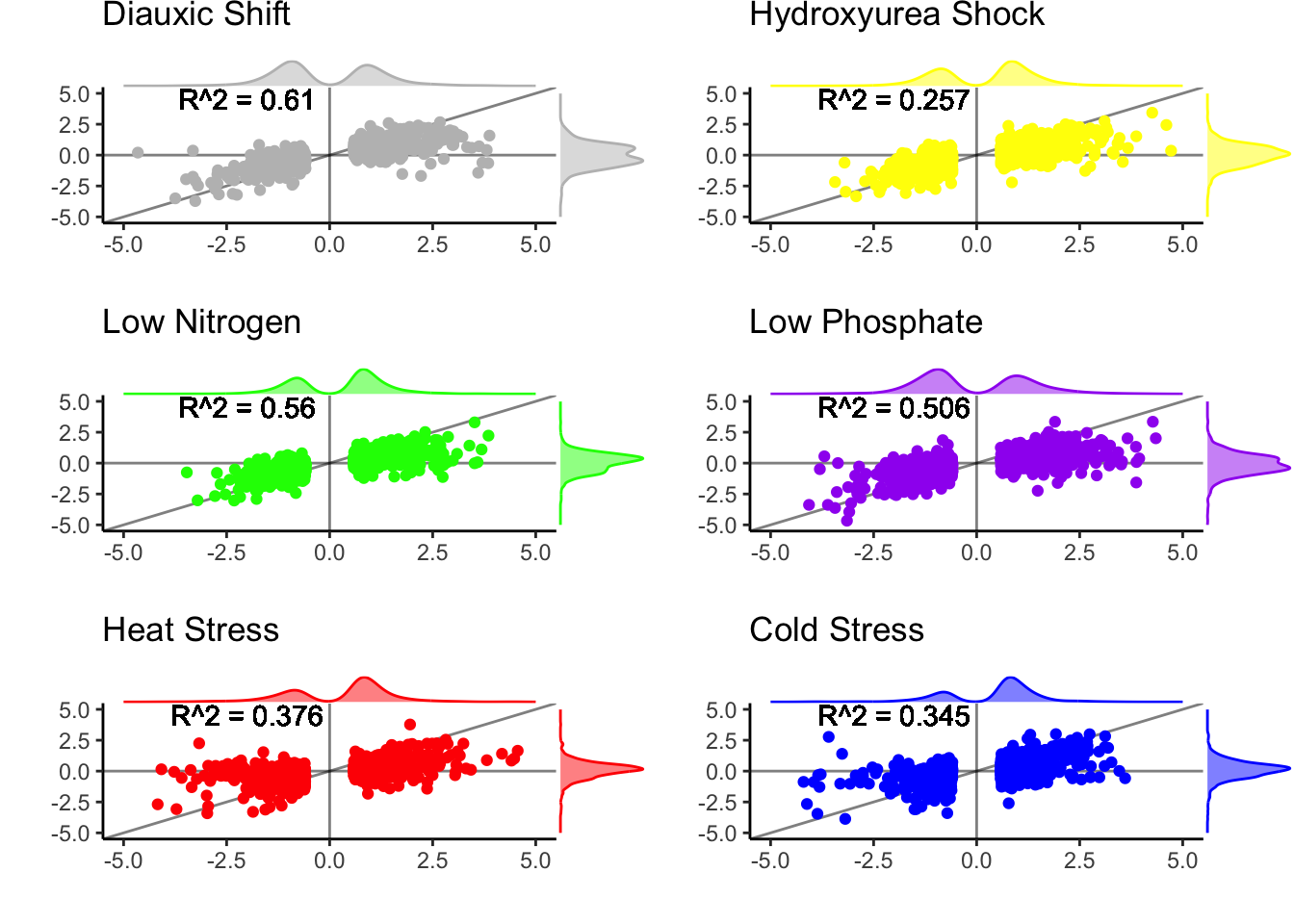
What better way to show this than that looking at the small fraction of genes that are diverging in level in trans the most—that are diverging in level in the parents and have the smallest log2 fold change between hybrid alleles.
# pct genes in "transest level" group for figure
plotdf <- finaldf |>
filter(level == "diverged") |>
mutate(is_trans_upcer = (effect_size_species > trans_lev_thresh) &
(abs(effect_size_allele) < eff_thresh),
is_trans_uppar = (effect_size_species < -trans_lev_thresh) &
(abs(effect_size_allele) < eff_thresh))
plotdf |> mutate(is_trans = is_trans_upcer | is_trans_uppar) |>
group_by(experiment) |>
summarise(pct_trans = sum(is_trans)/n(),
pct_rest = sum(!is_trans)/n())## # A tibble: 6 × 3
## experiment pct_trans pct_rest
## <chr> <dbl> <dbl>
## 1 CC 0.173 0.827
## 2 Cold 0.221 0.779
## 3 HAP4 0.148 0.852
## 4 Heat 0.236 0.764
## 5 LowN 0.135 0.865
## 6 LowPi 0.242 0.758pctdf <- plotdf |> mutate(is_trans = is_trans_upcer | is_trans_uppar) |>
group_by(experiment) |>
summarise(trans = sum(is_trans)/n(),
cis = sum(!is_trans)/n()) |>
pivot_longer(cols = c("cis", "trans"), names_to = "mode",
values_to = "percent")
p <- ggplot(pctdf, aes(x="", y=percent, fill=mode)) +
scale_fill_discrete(limits = c("cis", "trans"),
type = c("salmon", "aquamarine")) +
geom_bar(stat="identity", width=1) +
coord_polar("y", start=0) +
theme_classic() +
theme(axis.ticks = element_blank(),
axis.text = element_blank(),
legend.position = "bottom") +
xlab("") +
ylab("") +
facet_wrap(~factor(experiment, levels = ExperimentNames,
labels = LongExperimentNames), nrow = 2, ncol = 3)
pdf("paper_figures/CisTrans/level_pcts.pdf",
width = 8, height = 4)
p
dev.off()## quartz_off_screen
## 2p
Average expression of the cis and trans portion of level divergers, Diauxic Shift as example:
plotdf <- finaldf |>
filter(level == "diverged") |>
mutate(is_trans_upcer = (effect_size_species > trans_lev_thresh) &
(abs(effect_size_allele) < eff_thresh),
is_trans_uppar = (effect_size_species < -trans_lev_thresh) &
(abs(effect_size_allele) < eff_thresh))
plotdf |> mutate(is_trans = is_trans_upcer | is_trans_uppar) |>
group_by(experiment) |>
summarise(pct_trans = sum(is_trans)/n(),
pct_rest = sum(!is_trans)/n())## # A tibble: 6 × 3
## experiment pct_trans pct_rest
## <chr> <dbl> <dbl>
## 1 CC 0.173 0.827
## 2 Cold 0.221 0.779
## 3 HAP4 0.148 0.852
## 4 Heat 0.236 0.764
## 5 LowN 0.135 0.865
## 6 LowPi 0.242 0.758# upcer, trans
gene_idxs <- filter(plotdf, experiment == "HAP4" & cer == 1 &
par == 1 & is_trans_upcer) |>
select(gene_name) |> pull()
p <- annotate_figure(plotGenes(gene_idxs, .quartet = TRUE,
.experiment_name = "HAP4"),
top = paste(length(gene_idxs), "genes"))## `summarise()` has grouped output by 'group_id',
## 'gene_name', 'experiment'. You can override using the
## `.groups` argument.
## `summarise()` has grouped output by 'time_point_num',
## 'experiment'. You can override using the `.groups`
## argument.
## Adding missing grouping variables: `time_point_num`,
## `experiment`
## Adding missing grouping variables: `time_point_num`,
## `experiment`p
pdf("paper_figures/CisTrans/level_trans_HAP411_upcer.pdf", width = 3, height = 3)
p
dev.off()## quartz_off_screen
## 2# upcer, cis
gene_idxs <- filter(plotdf, experiment == "HAP4" & cer == 1 &
par == 1 & !is_trans_upcer & !is_trans_uppar &
effect_size_species > 0) |>
select(gene_name) |> pull()
p <- annotate_figure(plotGenes(gene_idxs, .quartet = TRUE,
.experiment_name = "HAP4"),
top = paste(length(gene_idxs), "genes"))## `summarise()` has grouped output by 'group_id',
## 'gene_name', 'experiment'. You can override using the
## `.groups` argument.
## `summarise()` has grouped output by 'time_point_num',
## 'experiment'. You can override using the `.groups`
## argument.
## Adding missing grouping variables: `time_point_num`,
## `experiment`
## Adding missing grouping variables: `time_point_num`,
## `experiment`p
pdf("paper_figures/CisTrans/level_cis_HAP411_upcer.pdf", width = 3, height = 3)
p
dev.off()## quartz_off_screen
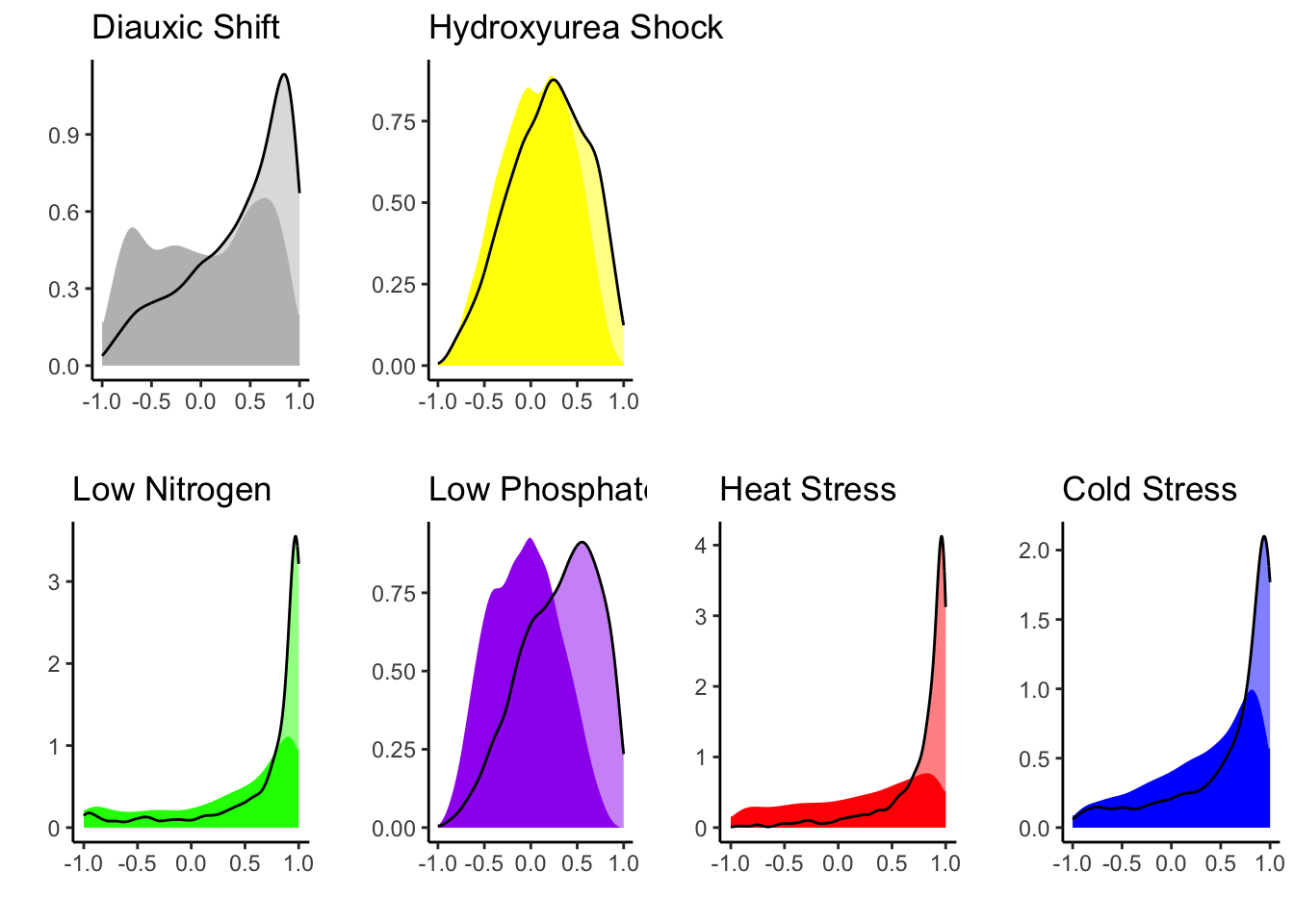
## 2Plasticity divergence tends to be in trans
plotdf <- finaldf |>
filter(plasticity == "diverged") |>
mutate(is_cis = cor_hybrid < cis_dyn_thresh)
plotdf |> group_by(experiment) |>
summarise(cis = sum(is_cis, na.rm = TRUE)/n(),
trans = sum(!is_cis, na.rm = TRUE)/n())## # A tibble: 6 × 3
## experiment cis trans
## <chr> <dbl> <dbl>
## 1 CC 0.161 0.838
## 2 Cold 0.103 0.896
## 3 HAP4 0.148 0.852
## 4 Heat 0.0321 0.967
## 5 LowN 0.0858 0.913
## 6 LowPi 0.105 0.893pctdf <- plotdf |> group_by(experiment) |>
summarise(cis = sum(is_cis, na.rm = TRUE)/n(),
trans = sum(!is_cis, na.rm = TRUE)/n()) |>
pivot_longer(cols = c("cis", "trans"), names_to = "mode",
values_to = "percent")
p <- ggplot(pctdf, aes(x="", y=percent, fill=mode)) +
scale_fill_discrete(limits = c("cis", "trans"),
type = c("salmon", "aquamarine")) +
geom_bar(stat="identity", width=1) +
coord_polar("y", start=0) +
theme_classic() +
theme(axis.ticks = element_blank(),
axis.text = element_blank(),
legend.position = "bottom") +
xlab("") +
ylab("") +
facet_wrap(~factor(experiment, levels = ExperimentNames,
labels = LongExperimentNames), nrow = 2, ncol = 3)
p
pdf("paper_figures/CisTrans/plasticity_pcts.pdf",
width = 8, height = 4)
p
dev.off()## quartz_off_screen
## 2Average expression of Diauxic Shift example:
plotdf <- finaldf |>
filter(plasticity == "diverged") |>
mutate(is_cis = cor_hybrid < cis_dyn_thresh)
plotdf |> group_by(experiment) |>
summarise(cis = sum(is_cis, na.rm = TRUE)/n(),
trans = sum(!is_cis, na.rm = TRUE)/n())## # A tibble: 6 × 3
## experiment cis trans
## <chr> <dbl> <dbl>
## 1 CC 0.161 0.838
## 2 Cold 0.103 0.896
## 3 HAP4 0.148 0.852
## 4 Heat 0.0321 0.967
## 5 LowN 0.0858 0.913
## 6 LowPi 0.105 0.893# cis
gene_idxs <- filter(plotdf, experiment == "HAP4" & cer == 2 &
par == 1 & is_cis) |>
select(gene_name) |> pull()
p <- annotate_figure(plotGenes(gene_idxs, .quartet = TRUE,
.experiment_name = "HAP4"),
top = paste(length(gene_idxs), "genes"))## `summarise()` has grouped output by 'group_id',
## 'gene_name', 'experiment'. You can override using the
## `.groups` argument.
## `summarise()` has grouped output by 'time_point_num',
## 'experiment'. You can override using the `.groups`
## argument.
## Adding missing grouping variables: `time_point_num`,
## `experiment`
## Adding missing grouping variables: `time_point_num`,
## `experiment`p
pdf("paper_figures/CisTrans/plasticity_cis_HAP421.pdf",
width = 3, height = 3)
p
dev.off()## quartz_off_screen
## 2# trans
gene_idxs <- filter(plotdf, experiment == "HAP4" & cer == 2 &
par == 1 & !is_cis) |>
select(gene_name) |> pull()
p <- annotate_figure(plotGenes(gene_idxs, .quartet = TRUE,
.experiment_name = "HAP4"),
top = paste(length(gene_idxs), "genes"))## `summarise()` has grouped output by 'group_id',
## 'gene_name', 'experiment'. You can override using the
## `.groups` argument.
## `summarise()` has grouped output by 'time_point_num',
## 'experiment'. You can override using the `.groups`
## argument.
## Adding missing grouping variables: `time_point_num`,
## `experiment`
## Adding missing grouping variables: `time_point_num`,
## `experiment`p
pdf("paper_figures/CisTrans/plasticity_trans_HAP421.pdf",
width = 3, height = 3)
p
dev.off()## quartz_off_screen
## 2